Complex natural systems (petroleum, bio-fuel, natural organic matter, eco-metabolites)
Natural organic matrices consist of numerous molecular compartments with diverse structural motifs. Ultra-high resolution mass spectrometry is the only analytical method which can describe such systems on a scale of individual molecules. This enables researchers to study global carbon cycles, the metabolome of bacterial communities; to predict properties of heavy petroleum fractions and develop nature-like functionalized organic and composite materials.
The typical Fourier transform mass-spectrum of complex mixtures consists of thousands of well-resolved monoisotopic peaks, which can be assigned by exact molecular formulae. Data can be plotted on conventional 2D diagrams, which facilitates a visual comparison of different samples. The advantage of FT MS is that the relative intensities of peaks are suitable for statistical assessment. This is essential for building classification and SAR models, which is the major goal of the project
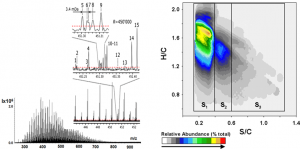 Fig. 1. Typical FTICR mass-spectrum of crude oil and the corresponding van Krevelen plot
Fig. 1. Typical FTICR mass-spectrum of crude oil and the corresponding van Krevelen plot
The limitation of FT MS analysis is tolerance to structural isomers. Molecular formula assigned to the peak may correspond to numerous amount of isomers. The project is also aimed to develop a strategy, which would extend chemical information on the individual components of complex mixtures obtained by mass spectrometry. Two approaches were suggested: selective chemical modification and data mining in big databases of chemical and biological properties.
The chemical modification should be rapid, mild, and result in the formation of ions, which are absent in the mass spectrum of the parent material. Stable isotopic labeling is the most promising approach. Several in-source and off-line exchange and labeling reactions have been already applied to humic substances, petroleum, dissolved organic matter, fungi metabolite, etc. Important information about their component structures was obtained by enumeration of reactive sites of ions. Separation techniques, as well as other labeling reactions, are under development to find out structures of individual components.
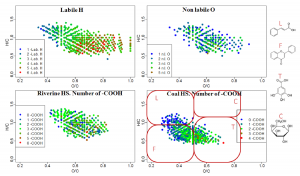 Fig. 2. Enumeration of specific atoms in individual components of natural organic matter
Fig. 2. Enumeration of specific atoms in individual components of natural organic matter
Another approach engages data mining of FTMS results for in silico search of possible structures in big databases, such as PubChem and ChEMBL. The appropriate candidates must possess similar chemical properties or specific biological activity. At the same time, structural fragments of compounds from databases must match the results of isotopic labeling. Combination of chemoinformatic and labeling approaches substantially narrows the number of possible isomers. The chemoinformatic approach includes standardization of FTMS data, application of different algorithms to assess molecular space, chemical similarity, and in silico search of a big array of data in databases. Application of most modern algorithms is required to predict chemical properties and biological activity of natural mixtures based on native and isotopic-exchange FTMS data.
Fig. 3. FTMS data-mining approach