PyFragMS
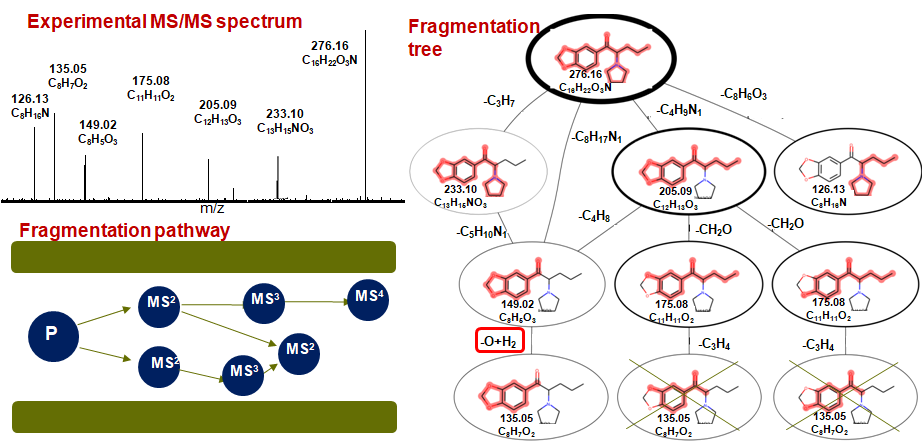
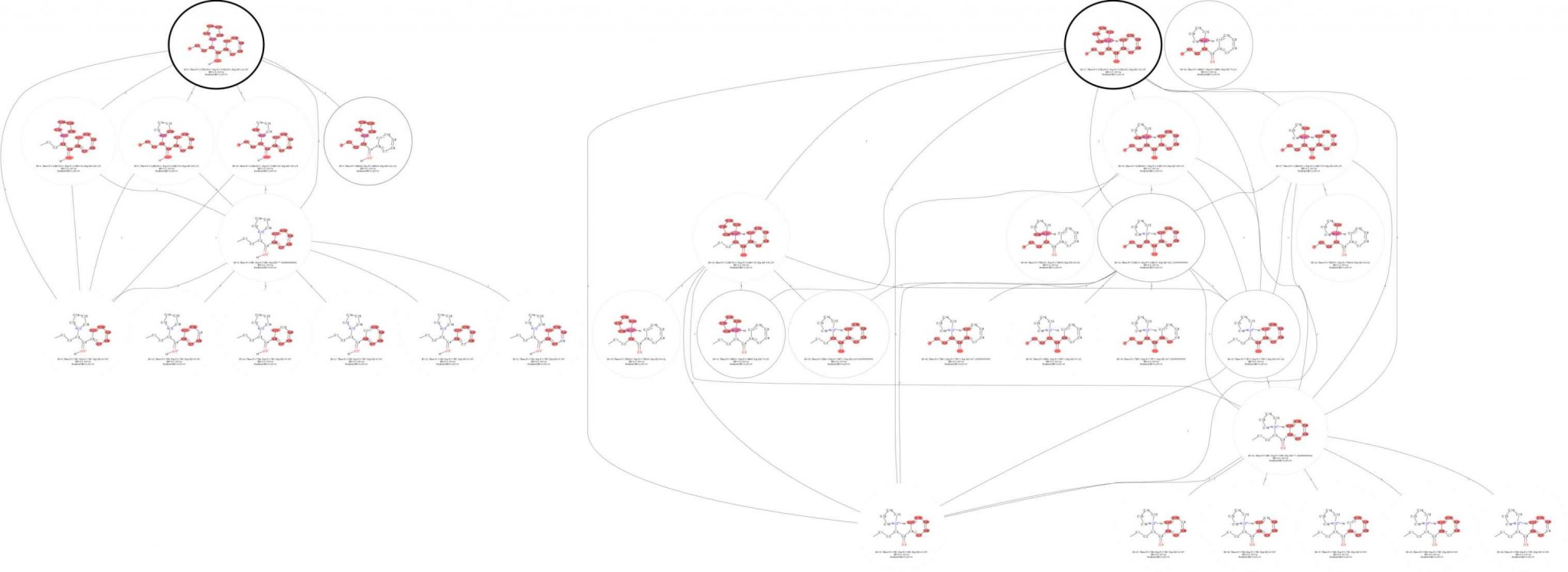
Dissociation induced by the accumulation of internal energy via collisions of ion with neutral molecules is one of the most important fragmentation techniques in mass spectrometry and the identification of small singly charged molecules is based mainly on the consideration of the fragmentation spectrum. Unfortunately, despite the growth of databases of experimentally measured MS/MS spectra (such as MzCloud, Metlin, etc.), numerous theoretical works and developing software for predicting MS/MS fragments in silico from SMILES patterns (such as MetFrag, CFM-ID, CSI:FingerID, etc.) the fragmentation mechanisms and pathways are not fully understood. Here we are proposing to use a combination of two powerful approaches: computing fragmentation trees that carry information of all consecutive fragmentations and consideration of the MS/MS data of isotopically labeled compounds. We have created a PyFragMS – a WEB tool consisting of database of annotated MS/MS spectra of isotopically labeled molecules (after H/D and 16O/18O exchange) and instruments for creating of the fragmentation tree for arbitrary molecule. Using PyFragMS we investigated how the site of protonation influence the fragmentation pathway for small molecules.
PyFragMS front-end was developed using Anwil (https://anvil.works/). Anvil allows to build web apps, using Python. PyFragMS back-end was developed using Python. Operations with molecules was realized using RDKit library (https://www.rdkit.org/). The PyFragMS WEB interface is described in Fig.1 (also see Supporting Information for more details). For plotting of fragmentation the GraphViz software was used.
PyFragMS database contains previously published MS/MS spectra of compounds after H/D exchange. And data acquired by us (MS/MS spectra of compounds after H/D and 16O/18O exchange). Assigning of molecular formulas to fragment ions was done using Sirius 4 software. Currently database contains spectra for 450 compounds for pos-ESI and 85 compounds for neg-ESI. For building fragmentation trees user can use data stored in database or upload his own data.
For the convenience of users we have recorded a video tutorial explaining the architecture of PyFragMS and step-by-step instruction how to use it. The tutorial is available at the PyFragMS website.
Link to PyFragMs:
https://pyfragms.anvil.app/
Link to video tutorial
Hope you will enjoy using PyFragMS! In case if something is not working or you have any questions, or wish to share your data to be included into database, please email us at (Yury Kostyukevich).